CMOST Plug-in (v1.0)
Table of Contents
- Introduction
- What is CMOST?
- CMOST Plugin
1.0 Introduction
The CMOST plug-in acts as a user interface for the CMOST database. It uses the DISCOVERYspace framework to allow the user to map experimental SAGE tags using the CMOST approach. It also draws upon the many other resources and features that DISCOVERYspace offers.
One of the most prominent problems currently with SAGE is the inability to assign annotations to many experimental SAGE tags (also known as “mapping tags”). Experimental SAGE tags that are “unmappable” do not tell the investigator anything about the expression of gene(s) that the tag is supposed to represent. Given that a significant proportion of experimental tags do not map to any sort of annotation using the current approaches, the usefulness of these unmappable tags are limited. The CMOST (Comprehensive Mapping of SAGE Tags) approach tries to alleviate this problem by providing a more, as the name states, comprehensive approach. More information about the CMOST approach is provided in the next section.
The CMOST plug-in is integrated into DISCOVERYspace in a way that may not be apparent at first. It is integrated into the menus of DISCOVERYspace. It does not have its own separate window. Usage of the plug-in will be shown in the following sections.
2.0 What is CMOST?
 |
Figure 1: General overview of CMOST
As stated in the introduction, current approaches to SAGE tag mapping leave a significant portion of tags unmapped. The CMOST approach alleviates this problem in a number of ways:
1. Data sources
CMOST draws tag data from 8 different data sources. These are:
1) MGC
2) RefSeq
3) Ensembl transcripts
4) Ensembl EST transcripts
5) Transcription units (based on Ensembl genes)
6) Golden path
7) Mitochondria (Genbank)
8) Non-protein coding genes (Genbank)
2. Sequence modification of tags
CMOST takes into account certain sequence modifications that a tag may have undergone. This may be attributed to experimental error, SNPs, indels, etc. It does this by simulating these anomalies through the modification of the tag sequence. There are 3 modifications that a tag goes through in CMOST. These are:
a) Single base permutation – A base in the tag (after the anchoring enzyme site), is exchanged with a A,C,T, and G (depending on the original base pair). Each tag produced has one modified base.
b) Single base insertion – A base position in the tag (after the anchoring enzyme site), is inserted with a A,C,T, or G each time. Each tag produced has one inserted base.
c) Single base deletion – A base in the tag (after the anchoring enzyme site), is deleted. Each tag produced has one deleted base.
3. Speed
Each SAGE library is run through CMOST prior to a user using those tag mappings. This ensures that all mapping data is already stored in the database. Therefore, any tags that a user wishes to map will receive the mapping results immediately without waiting for complex computations and sequence alignments to take place (given the tag has already been run through CMOST).
3.0 CMOST Plugin
The overall idea behind the CMOST plug-in has been designed to be fairly straightforward. The plug-in merely allows the user to establish a relationship either from an experimental SAGE tag to a data source, or vise versa.
3.1 Mapping SAGE Tags to Data sources with CMOST
3.1.1 CMOST Tag-to-Data Source Mapping Relationships
Generally, a user will start out with a list of experimental SAGE tags in the data viewer of DISCOVERYspace.
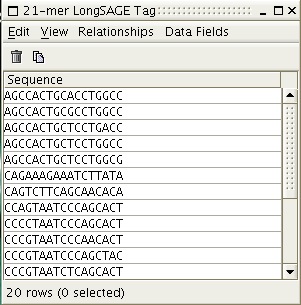
Figure 2: A list of experimental SAGE tags in DISCOVERYspace
By installing the CMOST plug-in, addition relationships are available to the user.
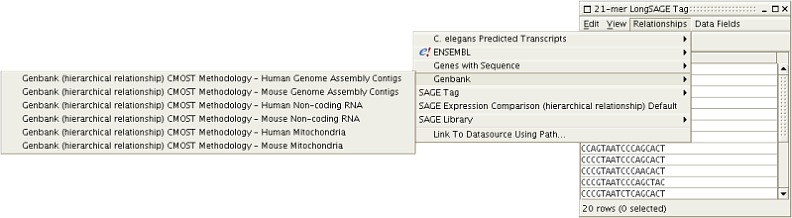
Figure 3: A few of the tag-to-data source relationships that CMOST adds
To map the experimental tags to a data source, the user selects the data source to map to. In this example the user has selected to map to the mouse genome assembly (Golden Path).
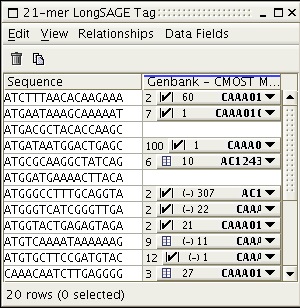
Figure 4: Results of CMOST mappings to the Mouse genome (Golden Path)
The user can choose which data sources he/she wishes the experimental SAGE tags to be mapped to.
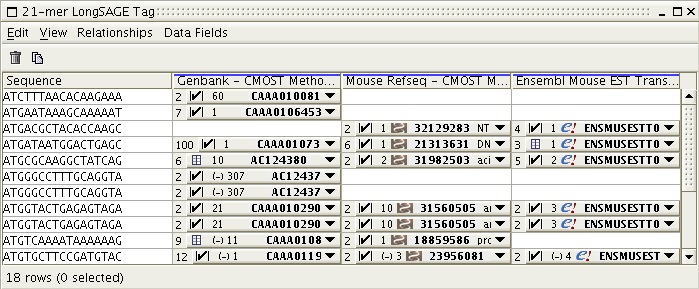
Figure 5: Results of CMOST mappings to various data sources
3.1.2 Description of CMOST Results
3.1.2.1 Successful Mapping Result
Each successful tag mapping is shown like the following:
![]()
Figure 6: A successful CMOST mapping result
There are 5 main sections to a tag mapping result:
Tag modification
![]() = Unmodified
= Unmodified
![]() = Single base permutation
= Single base permutation
![]() = Single base insertion
= Single base insertion
![]() = Single base deletion
= Single base deletion
Cut site and direction
The number represents the cut site number, starting with “1” as the 3'-most. If the mapping is antisense, then a “(-)” will appear before the site number.
Note: Currently, CMOST counts cut sites relative to the direction of the strand. So if there is an antisense mapping, the “(-) 1” means that it is the 3' most cut site relative to the antisense strand. This method of labeling cut sites is different from the method DISCOVERYspace uses and will be corrected in the future.
Data source
This icon is the visual representation of the data source that the tag has been mapped to.
Accession Number
This number is the accession number of the data source entry that the tag has been mapped to.
Annotation
This text is the description of the data source entry that the tag has been mapped to.
3.1.2.2 No-Mappings Result
If the SAGE tag mapped to none of the data sources, then a blank result is shown:
![]()
Figure 7: Unmappable CMOST Result
3.1.2.3 CMOST Mapping Not Available for Tag Result
If the SAGE tag has not been run through CMOST (either because the tag is from an external SAGE library, a new internal SAGE library, or from a SAGE library from a different species), then you will receive a “not available” result similar to the following:
![]()
Figure 8: The result returned from a tag that has not been run through the
CMOST pipeline
3.2 CMOST Best Mapping
(Sorry, the CMOST Best Mapping feature will be included in the next release which will be approximately 2-3 weeks after 3.1.4 is released. It is currently undergoing testing and further refinement.)
Click for screenshot
Figure 9: Screenshot of the best CMOST mapping being tested
3.3 CMOST Plug-in Parameters
CMOST Parameters are accessed by selecting on File -> Properties -> CMOST Plugin
3.3.1 CMOST PHP Layer URL
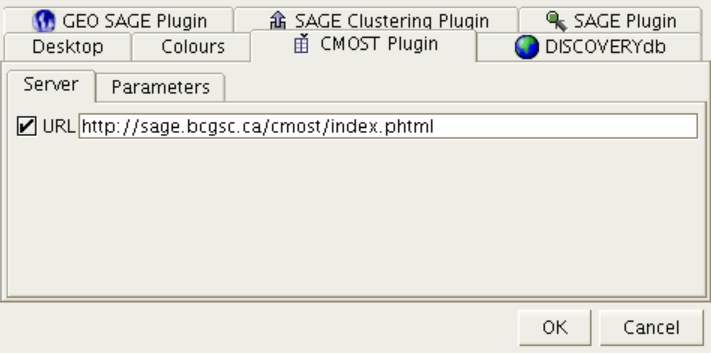
Figure 10: CMOST PHP Layer URL
The URL option shows the web address to the CMOST PHP layer. This usually should not be changed.
3.3.2 Maximum Number of Mappings to Return
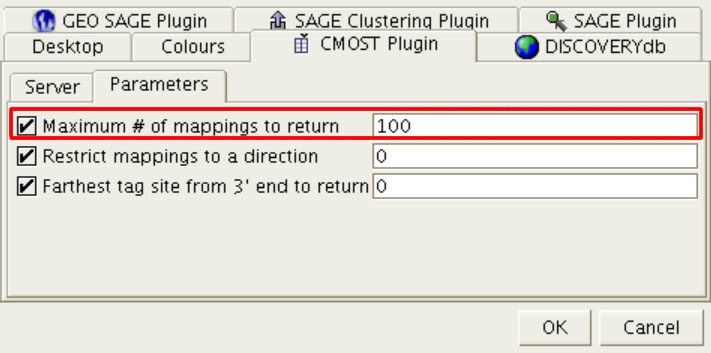
Figure 11: Maximum # of mappings to return parameter
The “Maximum # of mappings to return” option specifies the maximum number of mappings that the CMOST PHP layer will return to DISCOVERYspace. It is important to note that the greater the number of hits returned, the more burdened the system will be. The default is '100'.
3.3.3 Restrict Mappings to Sense or Antisense Only

Figure 12: Restrict mappings to a direction parameter
The “Restrict mappings to a direction” option specifies whether or not to restrict the mappings returned to a certain direction. The valid values are:
'0' - mappings to both directions will be returned.
'1' – mappings to the sense direction only will be returned.
'-1' – mappings to the antisense direction only will be returned.
The default is '0'.
3.3.4 Farthest Tag from 3’ End to Return
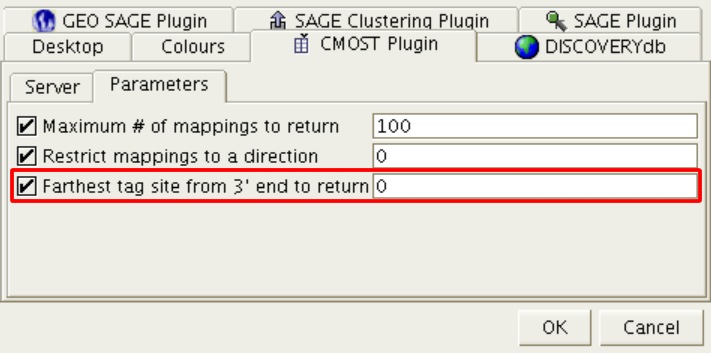
Figure 13: Farthest tag site from 3’-end to return parameter
The “Farthest tag site from 3' end to return” option specifies the highest anchoring enzyme site number that a hit has which will be returned. A value of '0' indicates that mappings to any site will be returned. The default is '0'.
DEREK LEUNG 16 FEB 2004 (Revision 1.0)