
A new Platform for Oncogenomic Reporting and Interpretation (PORI) integrates multiple knowledge bases into a program for sharing relevant data from genome analysis related to precision oncology.
Genomic data can be difficult to interpret. This is especially true when whole genomes and transcriptomes from tumour biopsies of advanced cancers are sequenced—as is the case in BC Cancer’s Personalized OncoGenomics (POG) program.
The process of comparing sequence data to reference genomes and publicly accessible knowledge bases is a big component of the POG program. Since the first patient was enrolled in 2012, the manual time required to review existing scientific literature for information on individual DNA sequence variants has been the most labour-intensive analysis step of the process.
”A lack of existing informatics tools needed to fully integrate whole genome and transcriptome data and the growing domain knowledge required us to start from scratch in building many of the methods, tools, and standards for the POG program,” says Caralyn Reisle, PhD student in the Jones lab.
Interpretation
This led to the creation of the Platform for Oncogenomic Reporting and Interpretation (PORI)—a collaborative effort by a multi-disciplinary team of clinicians, researchers, and software. Described in a paper published by Nature Communications, the interpretation portion of the platform is composed of a graph-based knowledge base called GraphKB.
 According to Caralyn, first author of the paper, GraphKB evolved from a spreadsheet to a proper databased to a graph model that could incorporate ambiguity and hierarchical relationships. “GraphKB stores information from literature and external databases in such a way that we can match variants found in a patient's tumour sample to the knowledge base in an automated manner, limiting the potential for manual mistakes.”
According to Caralyn, first author of the paper, GraphKB evolved from a spreadsheet to a proper databased to a graph model that could incorporate ambiguity and hierarchical relationships. “GraphKB stores information from literature and external databases in such a way that we can match variants found in a patient's tumour sample to the knowledge base in an automated manner, limiting the potential for manual mistakes.”
Reporting
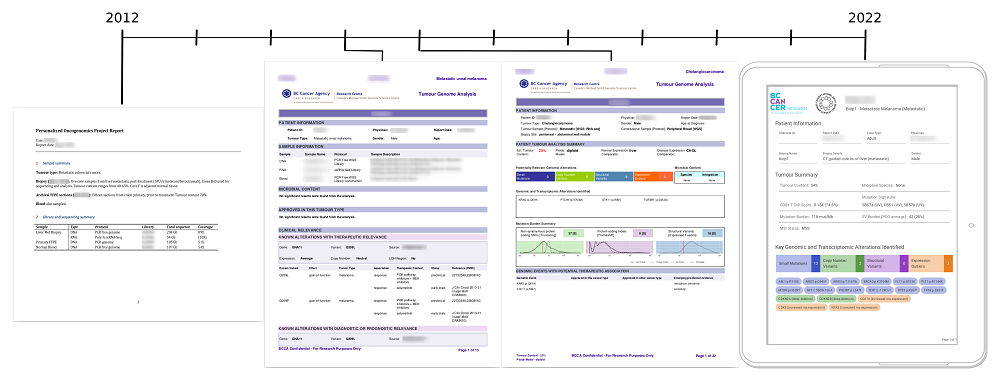
The other significant challenge for a program like POG is reporting results. Once clinically significant data is obtained, it must be communicated to both clinical and research experts in a comprehensive and efficient way so it can be incorporated into real-time treatment planning.
To accommodate this need, a reporting software—Integrated Pipeline Reports (IPR)—was incorporated into PORI. IPR generates genomic analysis results, including corresponding graphics, against a live version of GraphKB. It can be used to review and communicate data interactively through a web application or by the generation of PDF summary reports.
The PORI team held group meetings and incorporated feedback from user groups to make sure the reports were accessible and understandable. “One of the major outcomes of these design meetings was the need for both a condensed and extended version of the report,” says Caralyn.
Open Source
PORI is open source software, meaning that it is publicly accessible—anyone can see, modify, and distribute the code as needed. This was important to the team, who see it as an essential component of promoting reproducibility and transparency in research and healthcare, allowing both communities to evaluate, build on and learn from the software.
“A significant challenge to open-sourcing this platform was the sheer amount of code involved,” says Caralyn. “Through the release of PORI we have open sourced our entire reporting platform consisting of more than 10 code repositories worked on by more than a dozen developers.”
Future Directions
PORI represents an important first step in the creation of an open-sourced, production-ready, scalable software platform that can be used to standardize genomic data analysis and reporting. The team hopes it will be used by other sequencing centres and programs to aid in collaborative projects.
Although the platform is currently focused on creating research reports relevant to clinical trails, future iterations could include clinically accreditable formats of the report. Regardless of application, PORI will continue to improve as more information is added to GraphKB.
“As more knowledge is curated, each patient will match more annotations and reports will become increasingly verbose. Facilitating the complex analysis associated with precision oncology in cancer will not only have direct benefit to the patients analyzed but also the process as a whole through improved communication and transparency.”
Acknowledgements:
This work would not be possible without the participation of our patients and families, the POG team, the GSC platform, and the generous support of the BC Cancer Foundation and Genome British Columbia. Authors of the publication also acknowledge contributions towards equipment and infrastructure from Genome Canada and Genome BC, Canada Foundation for Innovation, the BC Knowledge Development Fund, and the Canada Research Chairs program to Dr. Steven Jones.
Learn More:
Read the first author’s Behind the Paper blog post for the Nature Portfolio Cancer Community and this interview from Science in Vancouver.
Learn more about POG. Watch this 5-mintue animation that provides an overview of the program in plain language, or this documentary on how the POG is applying precision to cancer research and treatment from CBC’s The Nature of Things.
Citation:
Reisle, C., Williamson, L.M., Pleasance, E., Davies, A., Pellegrini, B., Bleile, D.W., Mungall, K.L., Chuah, E., Jones, M.R., Ma, Y., Lewis, E., Beckie, I., Pham, D., Matiello Pletz, R., Muhammadzadeh, Al, Pierce, B.M., Li, J., Stevenson, R., Wong, H.m Bailey, L., Reisle, A., Douglas, M., Bonakdar, M., Nelson, J.M.T., Grisdale, C.J., Krzywinski, M., Fisic, A., Mitchell, T., Renouf, D.J., Yip, S., Laskin, J., Marra, M.A., Jones, S.J.M. A platform for oncogenomic reporting and interpretation. Nat Commun 13, 756 (2022). https://doi.org/10.1038/s41467-022-28348-y
*bold font indicates present and past members of the GSC.