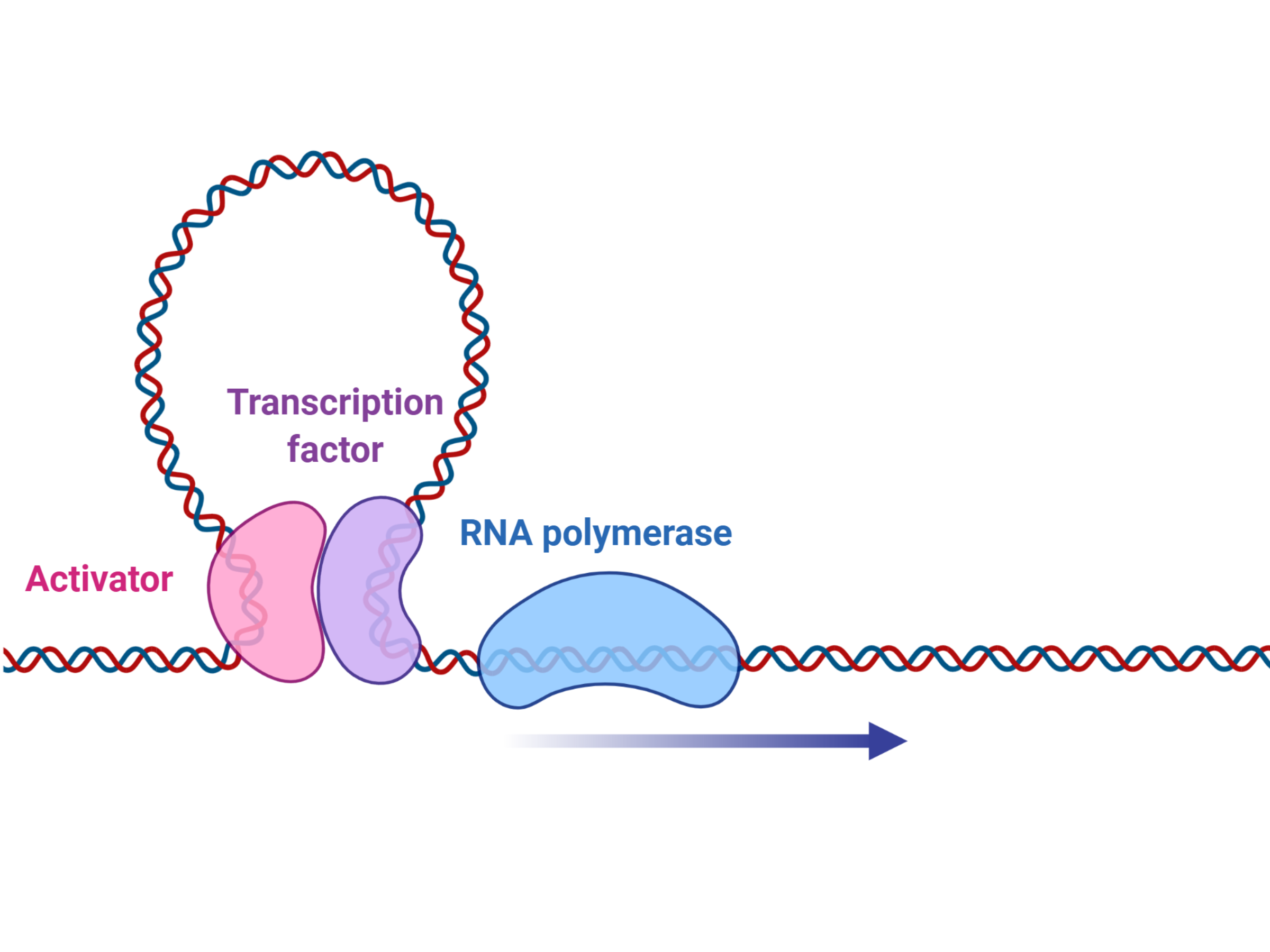
 The list of protein-coding drivers identified through sequencing in Diffuse Large B Cell Lymphoma (DLBCL) has begun to saturate and yet there remains a gap in our understanding of how these genetic alterations contribute to the transcriptional differences underlying known prognostic signatures and molecular subgroups. For example, dysregulation of NF-κB is the hallmark feature of ABC DLBCL but many ABC cases appear to lack the common driver mutations that are known to promote NF-κB. We are seeking driver mutations that contribute to some features of NHLs. We use data-driven comprehensive methods that allow identification of non-coding SSMs with a regulatory effect in cis or trans on genes/proteins relevant to malignancy.
The list of protein-coding drivers identified through sequencing in Diffuse Large B Cell Lymphoma (DLBCL) has begun to saturate and yet there remains a gap in our understanding of how these genetic alterations contribute to the transcriptional differences underlying known prognostic signatures and molecular subgroups. For example, dysregulation of NF-κB is the hallmark feature of ABC DLBCL but many ABC cases appear to lack the common driver mutations that are known to promote NF-κB. We are seeking driver mutations that contribute to some features of NHLs. We use data-driven comprehensive methods that allow identification of non-coding SSMs with a regulatory effect in cis or trans on genes/proteins relevant to malignancy.