Miscellaneous Analyses
Bruno Grande
2017-06-26
EBV Status
EBV status is currently determined based on the number of reads aligned to the EBV chromosome in GRCh38. Below is a plot showing the relationship between the EBV read ratio in DNA and RNA.
ebv_ratios %>%
filter(!is.na(rna_read_ratio)) %>%
mutate(
cohort = ifelse(grepl("^BL", patient), "BLGSP", "ICGC"),
dna_read_ratio = dna_read_ratio + 1e-6,
rna_read_ratio = rna_read_ratio + 1e-6) %>% {
ggplot(., aes(x = dna_read_ratio, y = rna_read_ratio, colour = cohort, label = patient)) +
geom_point() +
scale_x_log10() +
scale_y_log10() +
scale_colour_manual(values = colours$cohort) +
place_legend(c(0, 1)) +
labs(x = "EBV Read Ratio in DNA", y = "EBV Read Ratio in RNA", colour = "Cohort")}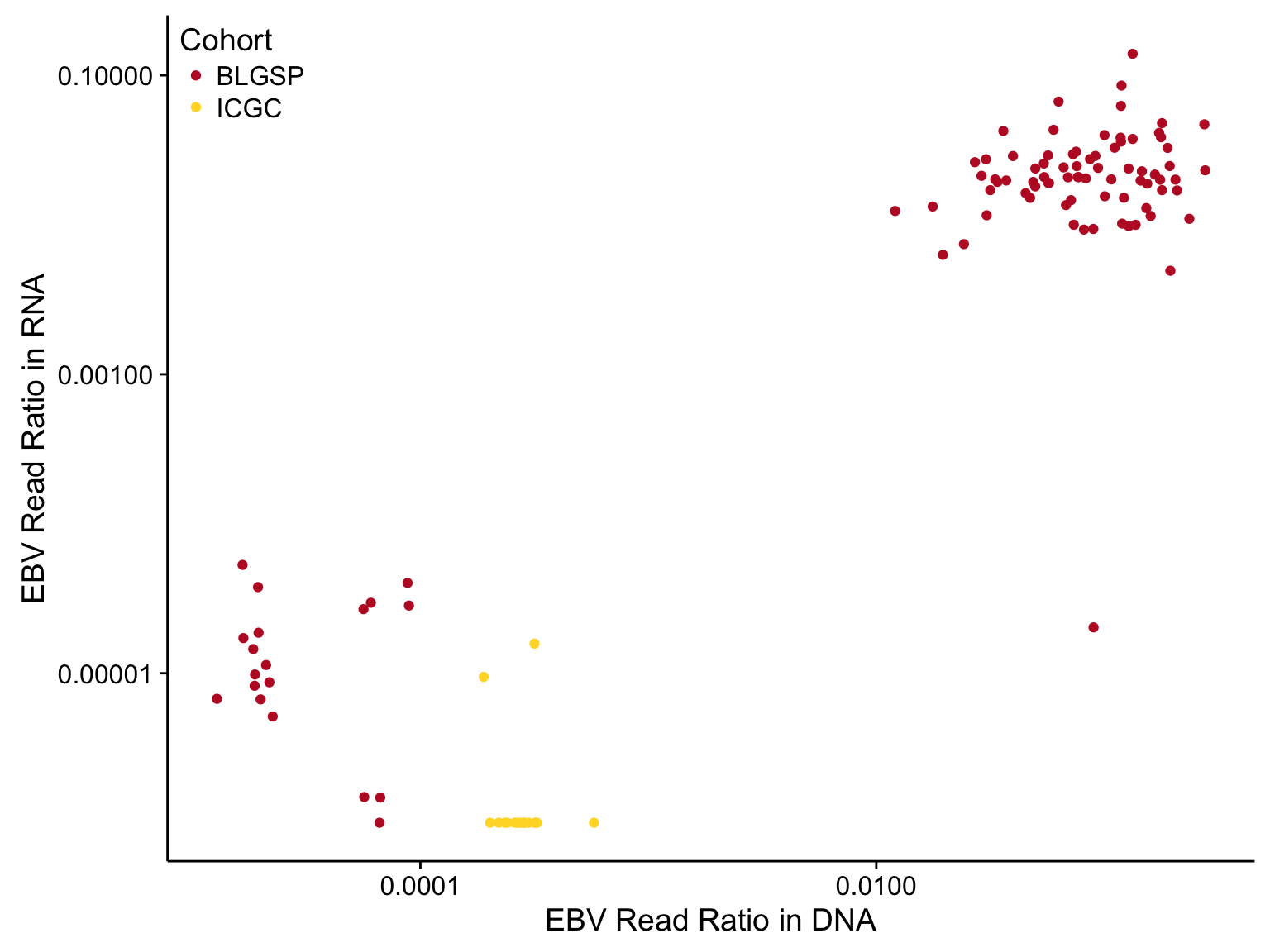
Tumour Content Estimates
metadata$genome %>%
select(patient, tumour_content, tumour_nuclei, ff_or_ffpe) %>%
drop_na() %>%
ggplot(aes(tumour_nuclei, tumour_content, colour = ff_or_ffpe)) +
geom_abline(slope = 1, intercept = 0, linetype = 2, colour = "grey") +
geom_point() +
coord_equal() +
scale_x_continuous(limits = c(0, 100)) +
scale_y_continuous(limits = c(0, 100)) +
scale_colour_manual(values = colours$ff_or_ffpe) +
place_legend(c(0, 1)) +
labs(title = "Percent Tumor Content",
x = "Pathologist estimate",
y = "Computational estimate",
colour = "FF/FFPE Status")
Centroblasts vs Centrocytes
cb_cc_degs <- get_sig_genes(salmon$clean$de$centro$lfc_0)
expr <- assay(salmon$clean$cvst)[cb_cc_degs,]
texpr <- expr[, grepl("^BL", colnames(expr))]
cbexpr <- expr[, grepl("^CB", colnames(expr))] %>% rowMedians()
ccexpr <- expr[, grepl("^CC", colnames(expr))] %>% rowMedians()
t_cb_cors <- cor(texpr, cbexpr)[,1]
t_cc_cors <- cor(texpr, ccexpr)[,1]
t_cors_df <- tibble(
patient = colnames(texpr),
Centroblasts = t_cb_cors,
Centrocytes = t_cc_cors)
t_cors_df %>%
gather(with, cor, -patient) %>%
ggplot(aes(x = with, y = cor, fill = with)) +
geom_boxplot() +
scale_fill_manual(values = colours$clinical_variant, breaks = NULL) +
labs(x = "Correlation with", y = "Pearson correlation")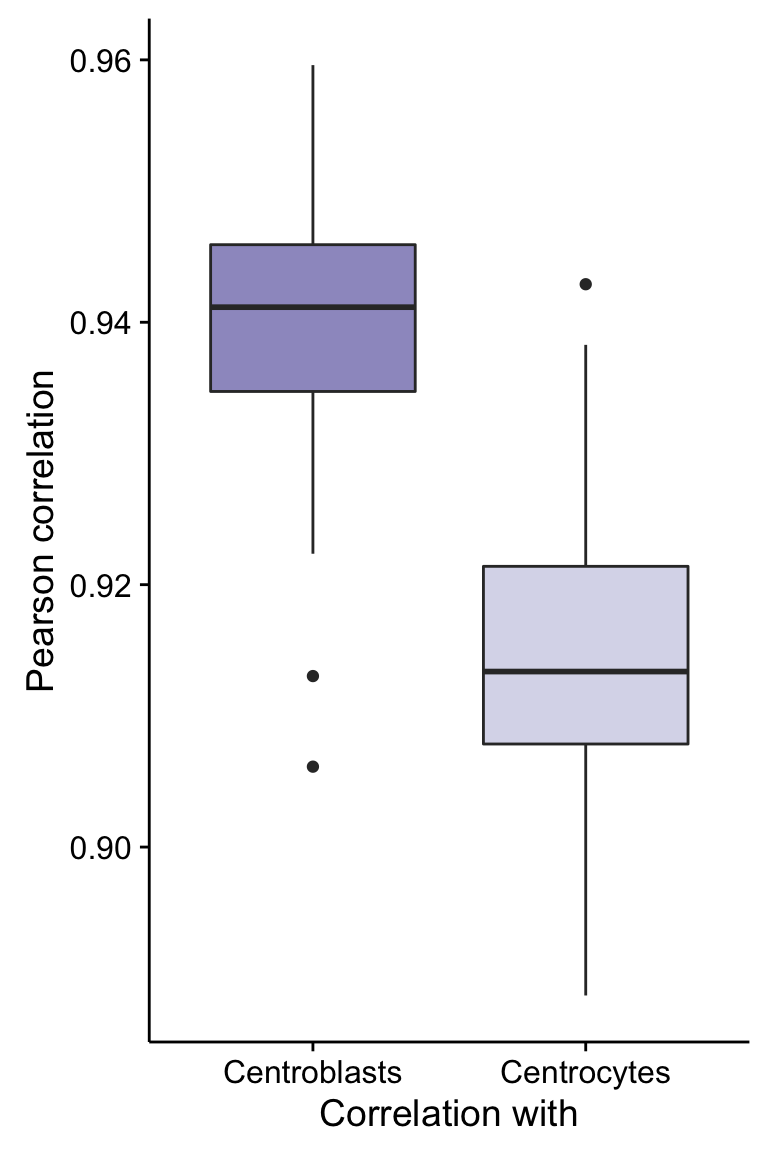
t_cors_df %>%
mutate(diff = Centroblasts / Centrocytes,
similar_to = ifelse(diff > 0, "Centroblasts", "Centrocytes")) %>%
ggplot(aes(x = 5, y = diff, colour = similar_to)) +
geom_hline(yintercept = 1, linetype = 2, colour = "grey") +
ggbeeswarm::geom_quasirandom(width = 0.01, method = "frowney") +
scale_x_continuous(labels = NULL) +
scale_y_continuous(limits = c(0.95, 1.05)) +
scale_colour_manual(values = colours$clinical_variant) +
place_legend(c(1,0)) +
labs(y = "Centroblasts / Centrocytes Pearson correlation ratio",
x = NULL, colour = "More similar to")
xCell
library(xCell)
xcell_vst <- xCellAnalysis(assay(salmon$clean$vst))[1] "Num. of genes: 10230"
Estimating ssGSEA scores for 489 gene sets.
|
| | 0%Using parallel with 4 cores
|
|= | 1%
|
|== | 2%
|
|=== | 3%
|
|=== | 4%
|
|==== | 5%
|
|===== | 6%
|
|====== | 7%
|
|======= | 8%
|
|======== | 9%
|
|========= | 10%
|
|========== | 11%
|
|========== | 12%
|
|=========== | 13%
|
|============ | 14%
|
|============= | 15%
|
|============== | 16%
|
|=============== | 17%
|
|================ | 17%
|
|================= | 18%
|
|================= | 19%
|
|================== | 20%
|
|=================== | 21%
|
|==================== | 22%
|
|===================== | 23%
|
|====================== | 24%
|
|======================= | 25%
|
|======================== | 26%
|
|======================== | 27%
|
|========================= | 28%
|
|========================== | 29%
|
|=========================== | 30%
|
|============================ | 31%
|
|============================= | 32%
|
|============================== | 33%
|
|=============================== | 34%
|
|=============================== | 35%
|
|================================ | 36%
|
|================================= | 37%
|
|================================== | 38%
|
|=================================== | 39%
|
|==================================== | 40%
|
|===================================== | 41%
|
|====================================== | 42%
|
|====================================== | 43%
|
|======================================= | 44%
|
|======================================== | 45%
|
|========================================= | 46%
|
|========================================== | 47%
|
|=========================================== | 48%
|
|============================================ | 49%
|
|============================================= | 50%
|
|============================================== | 51%
|
|=============================================== | 52%
|
|================================================ | 53%
|
|================================================= | 54%
|
|================================================== | 55%
|
|=================================================== | 56%
|
|==================================================== | 57%
|
|==================================================== | 58%
|
|===================================================== | 59%
|
|====================================================== | 60%
|
|======================================================= | 61%
|
|======================================================== | 62%
|
|========================================================= | 63%
|
|========================================================== | 64%
|
|=========================================================== | 65%
|
|=========================================================== | 66%
|
|============================================================ | 67%
|
|============================================================= | 68%
|
|============================================================== | 69%
|
|=============================================================== | 70%
|
|================================================================ | 71%
|
|================================================================= | 72%
|
|================================================================== | 73%
|
|================================================================== | 74%
|
|=================================================================== | 75%
|
|==================================================================== | 76%
|
|===================================================================== | 77%
|
|====================================================================== | 78%
|
|======================================================================= | 79%
|
|======================================================================== | 80%
|
|========================================================================= | 81%
|
|========================================================================= | 82%
|
|========================================================================== | 83%
|
|=========================================================================== | 83%
|
|============================================================================ | 84%
|
|============================================================================= | 85%
|
|============================================================================== | 86%
|
|=============================================================================== | 87%
|
|================================================================================ | 88%
|
|================================================================================ | 89%
|
|================================================================================= | 90%
|
|================================================================================== | 91%
|
|=================================================================================== | 92%
|
|==================================================================================== | 93%
|
|===================================================================================== | 94%
|
|====================================================================================== | 95%
|
|======================================================================================= | 96%
|
|======================================================================================= | 97%
|
|======================================================================================== | 98%
|
|========================================================================================= | 99%
|
|==========================================================================================| 100%heatmap_xcell_vst <-
xcell_vst %>%
most_var(15) %>%
plot_heatmap(colours, annotations[, -grep("lib_date", colnames(annotations))])
xcell_cvst <- xCellAnalysis(assay(salmon$clean$cvst))[1] "Num. of genes: 10230"
Estimating ssGSEA scores for 489 gene sets.
|
| | 0%Using parallel with 4 cores
|
|= | 1%
|
|== | 2%
|
|=== | 3%
|
|=== | 4%
|
|==== | 5%
|
|===== | 6%
|
|====== | 7%
|
|======= | 8%
|
|======== | 9%
|
|========= | 10%
|
|========== | 11%
|
|========== | 12%
|
|=========== | 13%
|
|============ | 14%
|
|============= | 15%
|
|============== | 16%
|
|=============== | 17%
|
|================ | 17%
|
|================= | 18%
|
|================= | 19%
|
|================== | 20%
|
|=================== | 21%
|
|==================== | 22%
|
|===================== | 23%
|
|====================== | 24%
|
|======================= | 25%
|
|======================== | 26%
|
|======================== | 27%
|
|========================= | 28%
|
|========================== | 29%
|
|=========================== | 30%
|
|============================ | 31%
|
|============================= | 32%
|
|============================== | 33%
|
|=============================== | 34%
|
|=============================== | 35%
|
|================================ | 36%
|
|================================= | 37%
|
|================================== | 38%
|
|=================================== | 39%
|
|==================================== | 40%
|
|===================================== | 41%
|
|====================================== | 42%
|
|====================================== | 43%
|
|======================================= | 44%
|
|======================================== | 45%
|
|========================================= | 46%
|
|========================================== | 47%
|
|=========================================== | 48%
|
|============================================ | 49%
|
|============================================= | 50%
|
|============================================== | 51%
|
|=============================================== | 52%
|
|================================================ | 53%
|
|================================================= | 54%
|
|================================================== | 55%
|
|=================================================== | 56%
|
|==================================================== | 57%
|
|==================================================== | 58%
|
|===================================================== | 59%
|
|====================================================== | 60%
|
|======================================================= | 61%
|
|======================================================== | 62%
|
|========================================================= | 63%
|
|========================================================== | 64%
|
|=========================================================== | 65%
|
|=========================================================== | 66%
|
|============================================================ | 67%
|
|============================================================= | 68%
|
|============================================================== | 69%
|
|=============================================================== | 70%
|
|================================================================ | 71%
|
|================================================================= | 72%
|
|================================================================== | 73%
|
|================================================================== | 74%
|
|=================================================================== | 75%
|
|==================================================================== | 76%
|
|===================================================================== | 77%
|
|====================================================================== | 78%
|
|======================================================================= | 79%
|
|======================================================================== | 80%
|
|========================================================================= | 81%
|
|========================================================================= | 82%
|
|========================================================================== | 83%
|
|=========================================================================== | 83%
|
|============================================================================ | 84%
|
|============================================================================= | 85%
|
|============================================================================== | 86%
|
|=============================================================================== | 87%
|
|================================================================================ | 88%
|
|================================================================================ | 89%
|
|================================================================================= | 90%
|
|================================================================================== | 91%
|
|=================================================================================== | 92%
|
|==================================================================================== | 93%
|
|===================================================================================== | 94%
|
|====================================================================================== | 95%
|
|======================================================================================= | 96%
|
|======================================================================================= | 97%
|
|======================================================================================== | 98%
|
|========================================================================================= | 99%
|
|==========================================================================================| 100%heatmap_xcell_cvst <-
xcell_cvst %>%
most_var(15) %>%
plot_heatmap(colours, annotations[, -grep("lib_date", colnames(annotations))])